Longitudinal hurdle models
Data
Recently, I have been modelling data that is longitudinal, contains excess zeros, and where the non-zero data is right-skewed and measured on a continuous scale, rather than being count data.
I’ll simulate a semi-realistic example data set from a lognormal hurdle model. The “random effects” for the pr(zero) and non-zero parts of the model are negatively correlated.
set.seed(180)
## 100 patients
id <- 1:100
## 2 timepoints
time <- c("1", "2")
## random effects
u <- rnorm(100, sd = 2)
v <- rnorm(100, mean = -0.95 * u, sd = sqrt((1 - 0.95^2) * 4)) # p(zero) is negatively correlated with Y*
## non-zero data (4 obs per id, at two timepoints)
ystar1 <- exp(rnorm(400, mean = u, sd = 1))
ystar2 <- exp(rnorm(400, mean = -0.5 + u, sd = 1))
## z = 1 if "cross hurdle", i.e. if not zero
z1 <- rbinom(400, size = 1, prob = 1 - plogis(-2 + v)) # p(cross hurdle) = 1 - p(zero)
z2 <- rbinom(400, size = 1, prob = 1 - plogis(-1 + v))
dat <- data.frame(y = c(z1 * ystar1, z2 * ystar2),
time = rep(time, each = 400),
id = rep(id, 8))In this data set there are 100 patients and two timepoints. For each patient, at each timepoint, I have simulated 4 independent observations (I’ve only done this to make model convergence a bit easier). The important point is that the data is correlated within patient, and also z (hurdle part) and ystar (non-zero part) are correlated, so that patients who start with a smaller (non-zero) y at the first timepoint are more likely to have y = 0 at the second timepoint. This can be clearly seen in the plot below.
library(tidyverse)
ggplot(data = dat,
mapping = aes(x = time, y = y, group = id)) +
geom_point() +
geom_line() +
scale_y_log10()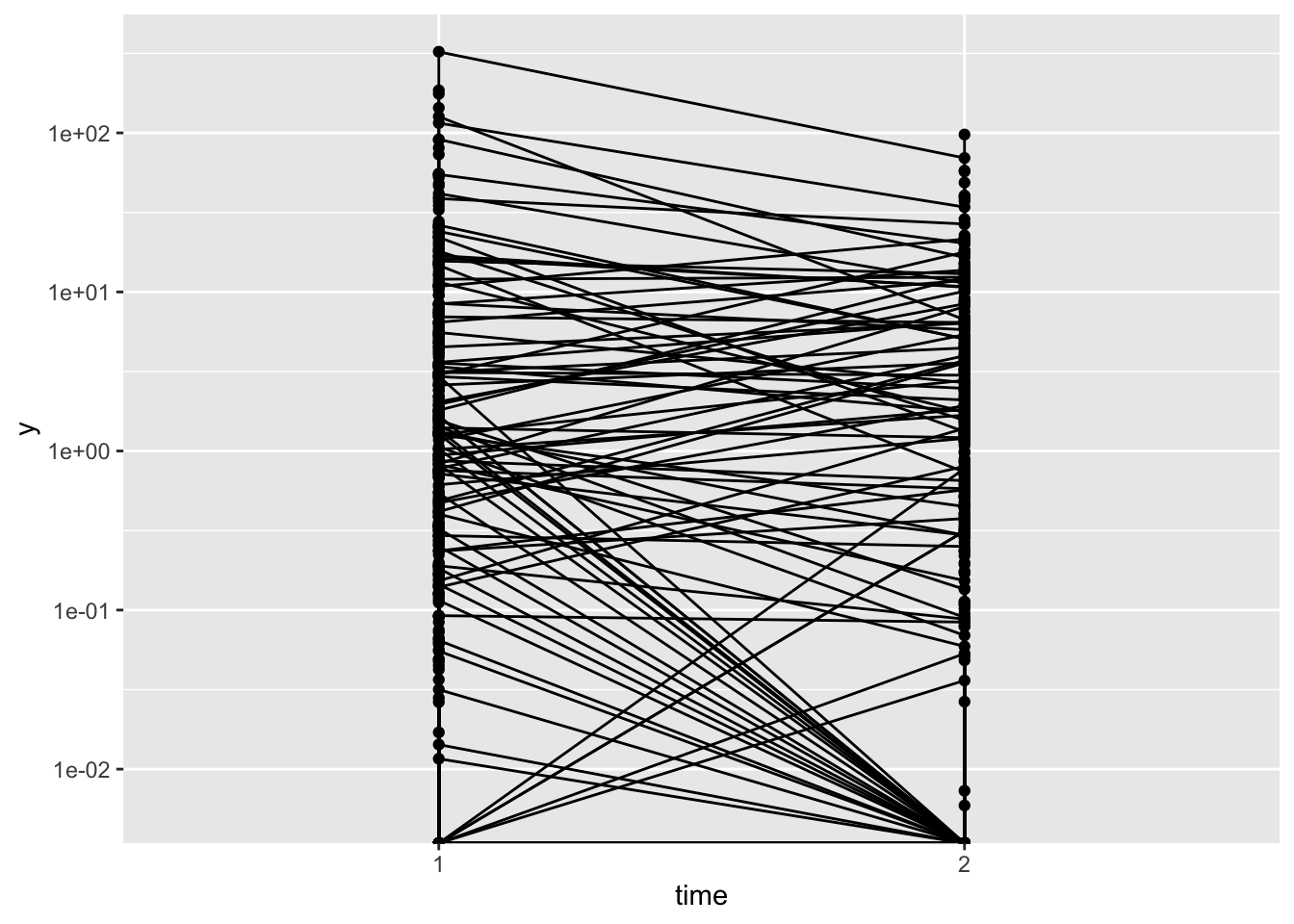
Fit the model
To fit the model I’m using the excellent brms package (https://github.com/paul-buerkner/brms)
Bürkner P. C. (2018). Advanced Bayesian Multilevel Modeling with the R Package brms. The R Journal. 10(1), 395-411. doi.org/10.32614/RJ-2018-017
library(brms)
fit_hurdle <- brm(bf(y ~ time + (1 | q | id),
hu ~ time + (1 | q | id)),
data = dat,
iter = 4000,
family = hurdle_lognormal(),
refresh = 0)summary(fit_hurdle)## Family: hurdle_lognormal
## Links: mu = identity; sigma = identity; hu = logit
## Formula: y ~ time + (1 | q | id)
## hu ~ time + (1 | q | id)
## Data: dat (Number of observations: 800)
## Samples: 4 chains, each with iter = 4000; warmup = 2000; thin = 1;
## total post-warmup samples = 8000
##
## Group-Level Effects:
## ~id (Number of levels: 100)
## Estimate Est.Error l-95% CI u-95% CI Rhat
## sd(Intercept) 1.96 0.16 1.69 2.29 1.00
## sd(hu_Intercept) 2.47 0.30 1.93 3.11 1.00
## cor(Intercept,hu_Intercept) -0.90 0.04 -0.97 -0.81 1.00
## Bulk_ESS Tail_ESS
## sd(Intercept) 1452 2688
## sd(hu_Intercept) 2979 5484
## cor(Intercept,hu_Intercept) 2539 4394
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept 0.07 0.20 -0.34 0.45 1.00 885 1970
## hu_Intercept -2.72 0.34 -3.43 -2.07 1.00 1662 3089
## time2 -0.33 0.08 -0.49 -0.18 1.00 14798 5985
## hu_time2 1.26 0.24 0.80 1.73 1.00 12452 5627
##
## Family Specific Parameters:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## sigma 0.94 0.03 0.88 1.00 1.00 11293 6175
##
## Samples were drawn using sampling(NUTS). For each parameter, Eff.Sample
## is a crude measure of effective sample size, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).Inference
From the output I can see that I have more-or-less recovered the parameters from my model. In practice, I could use this to make inference on the two parts of the model separately. In future posts I’ll discuss how to make inference/predictions when combining the two parts of the model.